What did scientists discover?
The so-called "KRAS gene" makes a protein (known just as "KRAS") that is involved in the regulation of cell division. Users have employed the MagLab's 21 tesla FT-ICR mass spectrometer to measure intact KRAS protein molecules, which produced the first clarified map of KRAS proteins in colon cancer tumors, including 28 previously unknown protein forms.
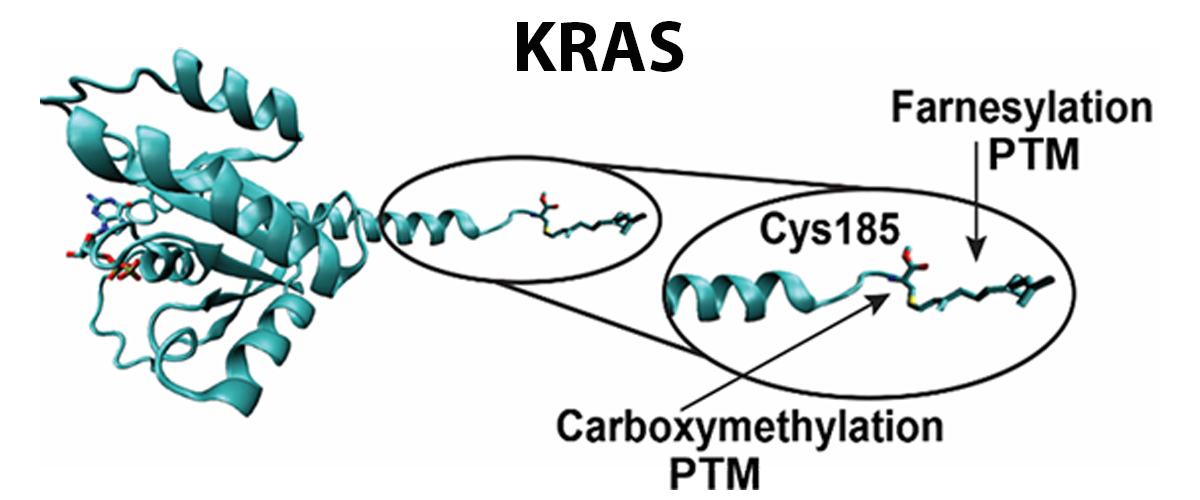
By measuring intact protein molecules, a new class of KRAS proteins were discovered that lack the terminal residue (C185*). With live cell imaging, the researchers found that the clipped KRAS did not bind to the cell membrane, where it serves as an on-off switch that regulates cell growth (Figure 1).
Why is this important?
Mutations in the KRAS gene are found in nearly 30% of human tumors. KRAS post-translational modification (PTM), which chemically changes the protein's structure, plays a crucial role in regulating the location of KRAS within the cell. This work may prove critically important for future development of anti-KRAS drugs that target membrane association in the battle against cancer.
Who did the research?
Lauren M. Adams1, Caroline J. DeHart2, Byron S. Drown1, Lissa C. Anderson3, William Bocik2, et al.
1Northwestern University; 2NCI-Frederick National Laboratory 3National MagLab
Why did they need the MagLab?
The MagLab's 21 T Ion Cyclotron Resonance magnet system provides exceptional mass resolving power and sensitivity, which dramatically improves detection of intact proteins. Before this work, 11 forms of KRAS were known. Twenty-eight additional forms were discovered here, revealing that that the KRAS protein landscape is far more complicated than previously thought.
Details for scientists
- View or download the expert-level Science Highlight, Mapping the KRAS Proteoform Landscape in Colorectal Cancer
- Read the full-length publication, Mapping the KRAS Proteoform Landscape in Colorectal Cancer Identifies Truncated KRAS4B that Decreases MAPK Signaling, in Journal of Biological Chemistry Data Set
Funding
This research was funded by the following grants:G.S. Boebinger (NSF DMR-2128556); Neil L. Kelleher (NIH P41 GM108569); Leidos Biomedical Research (HHSN261200800001E); and others
For more information, contact Chris Hendrickson.