What are the developments?
In this highlight, we feature Data Users working to develop software for improved identification of intact proteins by high resolution mass spectrometry (AKA - top-down proteomics). The original dataset was published in 2017 as a benchmark study on the performance of the 21T FT-ICR system for top-down proteomic analysis of colorectal cancer cells. It has since been cited in a poster and two papers for testing new data analysis algorithms and software packages, demonstrating the enhanced impact realized by of FAIR (Findable, Accessible, Interoperable, & Reusable) MagLab data.
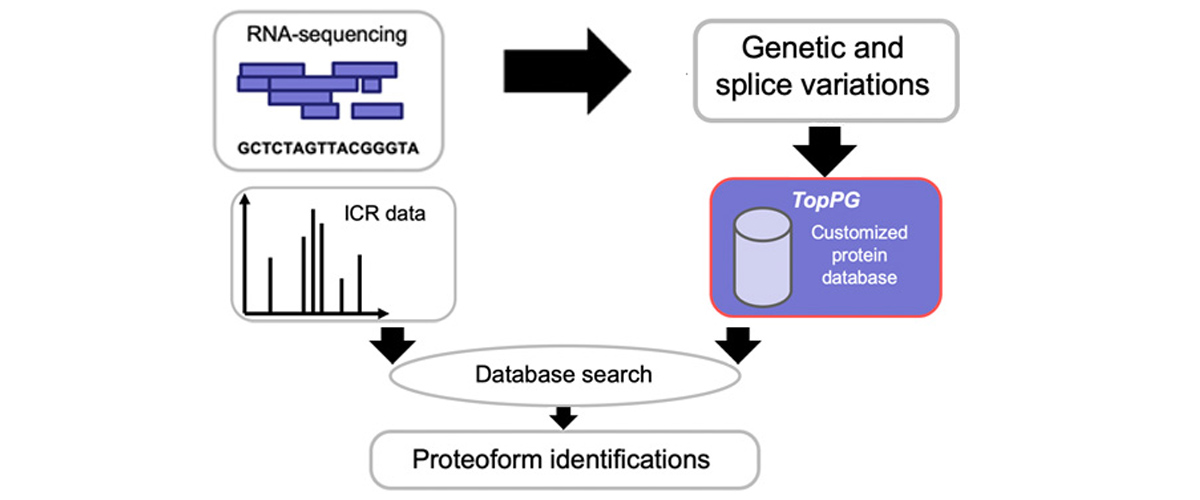
Researchers from multiple institutions independently harnessed data that had been previously collected at the MagLab by other researchers and stored under practices consistent with FAIR principles. The data was used to perform statistical analysis of fragmentation patterns to optimize search algorithms for identifying intact proteins from mass spectrometry data, demonstrate the discovery-mode workflow of MASH Explorer software, and test use of TopPG software to discover novel proteoforms involved in colorectal cancer.
Why is this important?
Reuse of the MagLab's Ion Cyclotron Resonance facility data improved understanding of protein fragmentation and aided the design and release of new algorithms and software tools. When the data were reanalyzed using databases created with TopPG, hundreds of previously unidentified proteoforms were discovered which might have direct clinical relevance to this colorectal cancer case. 'Data Users' demonstrate that MagLab data that is Findable, Accessible, Interoperable, and Reusable (FAIR) fosters knowledge, discovery, and innovation. As FAIR data practices grow, the impact of data generated at the MagLab will be amplified in a self-perpetuating cycle of new discoveries.
Why did they need the MagLab?
High quality data from intact proteins requires ultrahigh mass resolving power, mass accuracy, sensitivity, and spectral acquisition rate. The 21 T FT-ICR mass spectrometer provides all these capabilities, and this particular colorectal cancer dataset is gaining notoriety as a “gold standard” to test algorithm and software performance.
Impacted products of research
Ruixiang Sun, Ruimin Wang, Hao Chi, Chao Liu, Simin He, Ying Ge. TP 654: "Statistical Fragmentation Pattern Discovery of Intact Proteins Based on Their Large-scale Top-down MS/MS Spectra". 65th ASMS Conference on Mass Spectrometry and Allied Topics, Indianapolis, Indiana, June 4-8 (2017).
Zhijie Wu, David S. Roberts, Jake A. Melby, Kent Wenger, Molly Wetzel, Yiwen Gu, Sudharshanan Govindaraj Ramanathan, Elizabeth F. Bayne, Xiaowen Liu, Ruixiang Sun, Irene M. Ong, Sean J. McIlwain, and Ying Ge. "MASH Explorer: A Universal Software Environment for Top-Down Proteomics." Journal of Proteome Research. 2020, vol 19, pp 3867-3876. DOI: 10.1021/acs.jproteome.0c00469
Wenrong Chen and Xiowen Liu. "Proteoform Identification by Combining RNA-Seq and Top-Down Mass Spectrometry." Journal of Proteome Research. 2021, vol 20, pp 261-269. DOI: 10.1021/acs.jproteome.0c00369
Details for scientists
- View or download the expert-level Science Highlight, MagLab FAIR Data Empowers 'Data Users'
Funding
Creation of the original dataset was supported by the MagLab (G.S. Boebinger, NSF DMR-1644779, and the State of Florida) and by National Resource for Translational and Developmental Proteomics based at Northwestern University (N.L. Kelleher, NIH P41GM108569).
Data User research was funded by grants awarded to Ruixiang Sun1,3 (China '973' fund 2013CB911200, NSFC 31670837); Ying Ge2 (NIH R01GM125085, R01HL096971, GM117058, S10OD018475); Xiaowen Liu4 (NIH R01GM118470, R01GM125991, R01AI14625)
1Institute of Computing Technology, Chinese Academy of Sciences, Beijing, 2University of Wisconsin, Madison 3National Institute of Biological Sciences, Beijing, 4Indiana University—Purdue University Indianapolis; multiple departments
For more information, contact Christopher Hendrickson.